Fig. 4
Download original image
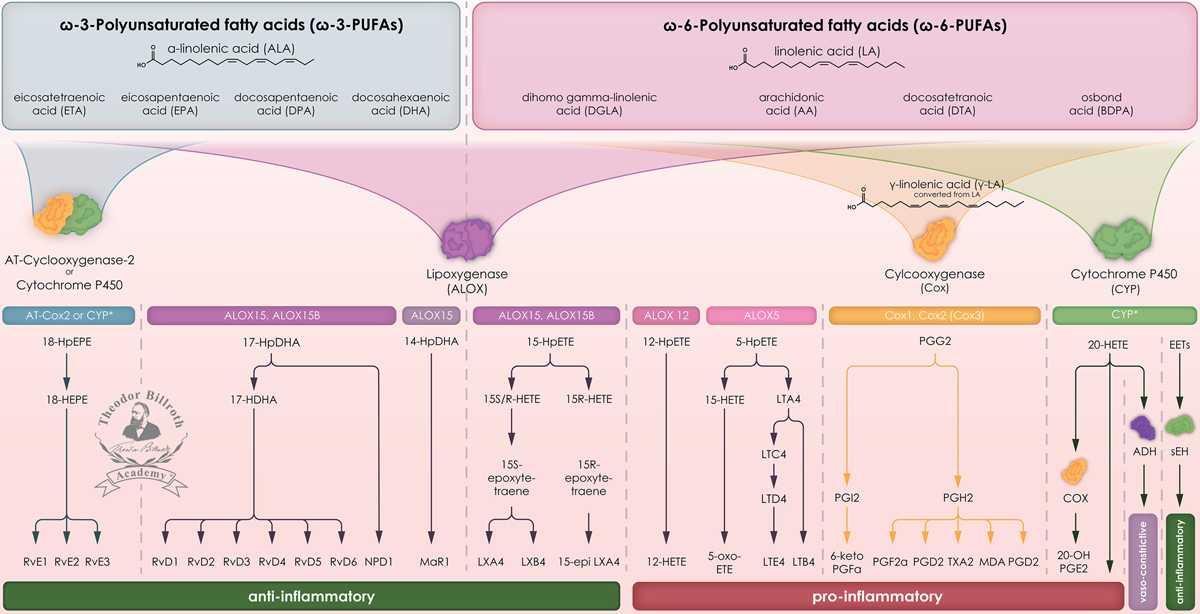
Eicosanoid metabolism and its relevance in inflammation. Nomenclature: Simplified scheme of the eicosanoid metabolism with formation from polyunsaturated fatty acids, including cyclooxygenase (Cox), lipoxygenase (ALOX), and cytochrome p450 (CYP) pathways with the formation of anti- and pro-inflammatory regulatory mediators. Common abbreviations are bold, followed by the common trivial names (if available) and (if available) by the name in accordance to the International Union of Pure and Applied Chemistry (IUPAC): ω-3-PUFAs N-3 polyunsaturated fatty acid; ALA α-linolenic acid, (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid; ETA eicosatetraenoic acid, all-cis-8,11,14,17-eicosatetraenoic acid; EPA eicosapentaenoic acid, (5Z,8Z,11Z,14Z,17Z)-eicosa- 5,8,11,14,17-pentenoic acid; DPA docosapentaenoic acid, 7,10,13,16,19-docosapentaenoic acid; DHA docosahexaenoic acid, (4Z,7Z,10Z,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoic acid; ω-6-PUFAs N-6 polyunsaturated fatty acid; LA linoleic acid, cis, cis-9,12-octadecadienoic acid; DGLA dihomo gamma-linolenic acid, (8Z,11Z,14Z)-8,11,14-Icosatrienoic acid; AA archidonic acid, (5Z,8Z,11Z,14Z)-5,8,11,14-eicosatetraenoic acid; DTA docosatetranoic acid, (7Z,10Z,13Z,16Z)-7,10,13,16-docosatetraenoic acid; BDPA osbond acid, (All-Z)-4,7,10,13,16-docosapentaenoic acid; AT-Cox2 aspirin-triggered cyclooxygenase 2; CYP* cytochrome P450 isoforms; 18-HpEPE 18-hydroxyeicosapentaenoic acid, 18-hydroxyicosa-2,4,6,8,10-pentaenoic acid; 18-HEPE (5Z,8Z,11Z,14Z,16E)-18-hydroxyicosa-5,8,11,14,16-pentaenoic acid; RvE1 resolvin E1, (5S,6Z,8E,10E,12R,14Z,16E,18R)-5,12,18-trihydroxyicosa-6,8,10,14,16-pentaenoic acid; RvE2 resolvin E2, (5S,6E,8Z,11Z,14Z,16E,18R)-5,18-dihydroxyicosa-6,8,11,14,16-pentaenoic acid; RvE3 resolvin E3, (5Z,8Z,11Z,13E,15E,18S)-17,18-dihydroxyicosa-5,8,11,13,15-pentaenoic acid; Cox cyclooxygenase; ALOX lipoxygenase, arachidonate lipoxygenase; ALOX15 15-lipoxygenase, 15-LOX, 15-LOX-1, arachidonate 15-lipoxygenase; ALOX15B 15-lipoxygenase type II, 15-LOX-2, arachidonate 15-lipoxygenase type B; ALOX12 12-lipoxygenase, 12-LOX, 12S-LOX, arachidonate 12-lipoxygenase 12S type; ALOX5 5- lipoxygenase, 5-LOX, arachidonate 5-lipoxygenase; 17-HpDHA 17S-hydroperoxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid; 17-HDHA 17S-hydroxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid; RvD1 resolvin D1, (4Z,7S,8R,9E,11E,13Z,15E,17S,19Z)-7,8,17-trihydroxydocosa-4,9,11,13,15,19-hexaenoic acid; RvD2 resolvin D2, (4Z,7S,8E,10Z,12E,14E,16R,17S,19Z)-7,16,17-trihydroxydocosa-4,8,10,12,14,19-hexaenoic acid; RvD3 resolvin D3, (4S,5E,7E,9E,13Z,15E,17R,19Z)-4,11,17-trihydroxydocosa-5,7,9,13,15,19-hexaenoic acid; RvD4 resolvin D4, (4S,6E,8E,10E,13E,15Z,17S,19Z)-4,5,17-trihydroxydocosa-6,8,10,13,15,19-hexaenoic acid; RvD5 resolvin D5, (5Z,7S,8E,10Z,13Z,15E,17S,19Z)-7,17-dihydroxydocosa-5,8,10,13,15,19-hexaenoic acid; RvD6 resolvin D6, (4S,5E,7Z,10Z,13Z,15E,17S,19Z)-4,17-dihydroxydocosa-5,7,10,13,15,19-hexaenoic acid; NPD1 neuroprotectin D1, protectin D1, (4Z,7Z,10R,11E,13E,15Z,17S,19Z)-10,17-dihydroxydocosa-4,7,11,13,15,19-hexaenoic acid; 14-HpDHA 14-hydro(peroxy)-docosahexaenoic acid, 4-[(1E,3E,5E,7E,9E)-nonadeca-1,3,5,7,9-pentaenyl]dioxete-3-carboxylic acid; MaR1 maresin 1, (4Z,7R,8E,10E,12Z,14S,16Z,19Z)-7,14-dihydroxydocosa-4,8,10,12,16,19-hexaenoic acid; 15-HpETE 15-hydroperoxy-eicosatetraenoic acid, (5Z,8Z,11Z,13E)-15-hydroperoxyicosa-5,8,11,13-tetraenoic acid; 15S/R-HETE 15S-HETE (5Z,8Z,11Z,13E,15S)-15-hydroxyicosa-5,8,11,13-tetraenoic acid; 15S-epoxy-tetraene 5,6-15S-HETE, 4-[(2S,3S)-3-[(1E,3E,5Z,7E,9S)-9-hydroxytetradeca-1,3,5,7-tetraenyl]oxiran-2-yl]butanoic acid; 15R-epoxy-tetraene lacks a bit of the activity attributed to their S stereoisomers of 15S-epoxy-tetraene; 15R-epoxy-tetraene lacks a bit of the activity attributed to their S stereoisomers of 15S-epoxy-tetraene; LXA4 lipoxin A4, (5S,6R,7E,9E,11Z,13E,15S)-5,6,15-trihydroxyicosa-7,9,11,13-tetraenoic acid; LXB4 lipoxin B4, (5S,6E,8Z,10E,12E,14R,15S)-5,14,15-trihydroxyicosa-6,8,10,12-tetraenoic acid; 15-epi-LXA4 (5R,6R,7E,9E,11Z,13E,15R)-5,6,15-trihydroxyicosa-7,9,11,13-tetraenoic acid; 12-HpETE 12-hydroperoxyicosa-5,8,10,14-tetraenoic acid; 12-HETE (5E,8Z,10Z,14Z)-12-hydroxyicosa-5,8,10,14-tetraenoic acid; 5-HpETE (6E,8Z,11Z,14Z)-5-hydroperoxyicosa-6,8,11,14-tetraenoic acid; 15-HETE (5Z,8Z,11Z,13E)-15-hydroxyicosa-5,8,11,13-tetraenoic acid; 5-oxo-ETE (6E,8Z,11Z,14Z)-5-oxoicosa-6,8,11,14-tetraenoic acid; LTA4 leukotriene A4, 4-[(2S,3S)-3-[(1E,3E,5Z,8Z)-tetradeca-1,3,5,8-tetraenyl]oxiran-2-yl]butanoic acid; LTC4 leukotriene C4, (5S,6R,7E,9E,11Z,14Z)-6-[(2R)-2-[[(4S)-4-amino-4-carboxybutanoyl]amino]-3-(carboxymethylamino)-3-oxopropyl]sulfanyl-5-hydroxyicosa-7,9,11,14-tetraenoic acid; LTD4 leukotriene D4, (5S,6R,7E,9E,11Z,14Z)-6-[(2R)-2-amino-3-(carboxymethylamino)-3-oxopropyl]sulfanyl-5-hydroxyicosa-7,9,11,14-tetraenoic acid; LTE4 leukotriene E4, (5S,6R,7E,9E,11Z,14Z)-6-[(2R)-2-amino-2-carboxyethyl]sulfanyl-5-hydroxyicosa-7,9,11,14-tetraenoic acid; LTB4 leukotriene B4, (5S,6Z,8E,10E,12R,14Z)-5,12-dihydroxyicosa-6,8,10,14-tetraenoic acid; LA linoleic acid, cis, cis-9,12-octadecadienoic acid; LA is not a direct substrate of prostaglandins − this occurs after metabolism to γ-LA; therefore LA here is in brackets; γ-LA gamma linolenic acid; Cox-1 cyclooxygenase 1; Cox-2 cyclooxygenase 2; Cox-3 splice variant and isoform of Cox-2; (therefore in brackets); PGG2 prostaglandin G2, (Z)-7-[(1S,4R,5R,6R)-5-[(E,3S)-3-hydroperoxyoct-1-enyl]-2,3-dioxabicyclo[2.2.1]heptan-6-yl]hept-5-enoic acid; PGI2 prostaglandin I2, prostacyclin I2, (5Z)-5-[(3aR,4R,5R,6aS)-5-hydroxy-4-[(E,3S)-3-hydroxyoct-1-enyl]-3,3a,4,5,6,6a-hexahydrocyclopenta[b]furan-2-ylidene]pentanoic acid; 6-keto-PGF1α 6-keto-prostaglandin F1alpha, 7-[(1R,2R,3R,5S)-3,5-dihydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]cyclopentyl]-6-oxoheptanoic acid; PGH2 prostaglandin H2, (Z)-7-[(1S,4R,5R,6R)-5-[(E,3S)-3-hydroxyoct-1-enyl]-2,3-dioxabicyclo[2.2.1]heptan-6-yl]hept-5-enoic acid; PGFF2α prostaglandine F2 alpha, (Z)-7-[(1R,2R,3R,5S)-3,5-dihydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]cyclopentyl]hept-5-enoic acid; PGD2 prostaglandin D2, (Z)-7-[(1R,2R,5S)-5-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-3-oxocyclopentyl]hept-5-enoic acid; PGE2 prostaglandin E2, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxocyclopentyl]hept-5-enoic acid; TXA2 thromboxane A2, (Z)-7-[(1S,2S,3R,5S)-3-[(E,3S)-3-hydroxyoct-1-enyl]-4,6-dioxabicyclo[3.1.1]heptan-2-yl]hept-5-enoic acid; MDA malondialdehyde, propanedial; Cox cyclooxygenase; Cox cyclooxygenase; 20-HETE 20-hydroxyeicosatetraenoic acid, (5Z,8Z,11Z,14Z)-20-hydroxyicosa-5,8,11,14-tetraenoic acid; 20-OH-PGE2 20-hydroxy prostaglandin E2; ADH alcohol dehydrogenase; 20-COOH-HETE 20-carboxy-hydroxyeicosatetraenoic acid; EETs epoxyeicosatrienoic acid; sEH soluble epoxide hydrolase; DHETs dihydroxyeicosatrienoic acids.

