| Issue |
4open
Volume 5, 2022
COVID-19 Articles
|
|
|---|---|---|
| Article Number | 16 | |
| Number of page(s) | 12 | |
| Section | Life Sciences - Medicine | |
| DOI | https://doi.org/10.1051/fopen/2022017 | |
| Published online | 12 August 2022 | |
Review Article
An update of serial interval estimates for COVID-19: a meta-analysis
Centre Hospitalier Universitaire de Lille, Département d’Information Médicale, 2 avenue Oscar Lambret, 59037 Lille Cedex, France
* Corresponding author: jean-francois.jusot@chu-lille.fr
Received:
4
April
2022
Accepted:
15
July
2022
Background: Serial interval (SI) is one of the most important parameter for COVID-19 modelling purposes as it is related to the reproduction rate of the infection. The first meta-analysis of serial interval were performed with a range of uncertainty in the estimate. This meta-analysis aimed to reduce the uncertainty estimates by assessing publications over a longer period. Methods: A literature search was performed for articles published between 1st December 2019 and 15th February 2022. It retrieved 117 eligible studies containing some 80 for 90 serial interval estimates. A random effects model was used. Heterogeneity was checked. To detect a publication bias, a funnel plot was performed using an Egger’s test. Results: For alpha variant, the serial interval was estimated at 5.17 days (95% CI = 4.87 – 5.47) with a significant heterogeneity (I2 = 97.1%). The meta-analysis did not exhibit evident publication bias (Egger’s test = −0.55, p = 0.58). The meta-analysis allowed for reducing uncertainty in estimating the serial interval, although subgroup analysis did not reduce it sufficiently and showed that studies using a gamma distribution of serial intervals exhibited the highest estimate of 5.6 days. Compared to the other variants of concern, alpha serial interval estimate was bigger than delta, 4.07 days, and omicron, 3.06 days. Conclusion: The meta-analysis was carried out as a real-time monitoring of this parameter to make a choice and a rapid assessment of the control measures implemented, and the effectiveness of the vaccination campaign. The meta-analysis was unable to provide a suitable estimate of serial intervals for COVID-19 modelling purposes although its uncertainty was reduced. Furthermore, serial intervals estimate for alpha variant was close to earlier reports and lower than previous publications, respectively. Another limitation is, that meta-analysis of COVID pandemic studies in principle contains and produces itself a significant source of heterogeneity.
Key words: Coronavirus / COVID-19 / Meta-analysis / Modelling / Contact tracing / Pandemic / Reproduction rate / SARS-CoV-2 / Statistics / Virus
© J.-F. Jusot et al., Published by EDP Sciences, 2022
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
One of the threats of the Coronavirus disease 2019 (COVID-19) pandemic is the occurrence of multiple variants since the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019. Among these variants, three are of public health concern, alpha, delta, and omicron, and these have contributed to huge pandemics [1, 2]. For this reason, the effectiveness of vaccines remains a relevant question and non-pharmaceuticals interventions should be ongoing. Furthermore, control of the pandemic remains a major public health challenge three years after the emergence of SARS-CoV-2.
Modelling helps to manage pandemics by understanding how an infectious disease spreads in the population, the choice and the assessment of adequate control strategies, mainly non-pharmaceutical interventions as is the case with the COVID-19 [3, 4]. At the onset of the COVID-19 pandemic, one of the priorities was to estimate essential parameters which allowed models to reliably describe the epidemic as it progressed. Among them, three were of great concern: the basic reproduction rate R0, the case fatality rate of the disease and the average duration of infection [5, 6] The basic reproduction rate is defined as the average number of secondary cases infected by a primary case during its infectious period in a population of fully susceptible subjects. Its value indicates whether the epidemic is able to continue (R0 > 1) or not in the population (R0 < 1 means the temporary decline or not of the epidemic). The level of uncertainty in the choice of a combination of control strategies is related to that in estimating model parameters, especially R0. This last is also a parameter of concern as it is used to estimate vaccine coverage to stop the transmission and/or its required effectiveness E through the formula (1 − 1/ R0)/E. As R0 is measured at the beginning of the epidemic, the effective reproduction number Rt describes its evolution.
R0 was initially estimated with a median value of 2.8 based on a review by Liu et al. [7]. The average duration of infection is another significant parameter to be considered. It is approximated by the serial interval (SI) or the generation time (GT). SI is defined as the difference between the date of onset of symptoms of a secondary case and an index case, while the GT corresponds to the difference between the dates of the onset of the infectious periods. As the onset of infectiousness is almost impossible to determine, SI is preferred to GT. The SI has been used in several analyses to model the spatiotemporal spread of the COVID-19 epidemic or to compare the effectiveness of control measures [3, 8–11]. Usually, reproduction rate values correlate positively with SI values. Nevertheless, negative SI can be observed if transmission to the secondary case occurs during the pre-symptomatic period of the primary case and the incubation time in the secondary case is shorter than that of the primary case. A SI distribution with a high variance taking into account negative values can skew this relationship. Higher variance of SI could explain an underestimation of reproduction rates [12, 13]. Moreover, distribution of the SI could bias the central estimation of the Rt by as much as 20–25% [14].
For COVID-19, meta-analyses of SI have been published in the first semester of 2020. They estimated the SI to be more than 5 days with varying confidence interval limits between 4.2 and 6.7 days [15–17]. The aim of this study is to provide an estimate of SI using a meta-analysis to produce more reliable reproduction rate for use in modelling.
Material and methods
Literature search
The search was performed for COVID-19 related articles published between 1st December 2019 and 15 February 2022. It was done from PubMed using the following keywords and combination: (serial AND interval) AND (SARS-CoV-2 OR COVID-19). The references of the publications found, in particular those of the meta-analyses or reviews, were also reviewed and potentially included. One serial interval (SI) estimate was kept if two publications or more from the same authors reported the same SI, standard deviation and number of infector/infected pairs.
Eligibility and exclusion criteria
All types of studies were searched, original articles as well as letters, brief communications, preprint, abstracts. Recorded were the type of publication, country, source population of cases, type of variant, starting date of the study, estimates of SI with negative values or not, SI statistical distribution, if SI estimates came from published data or could be re-estimated from raw data.
The following publications were excluded:
When one of the three parameters, mean, standard-deviation, number of infector/infected pairs, was/were not found in the publication and raw data was not available.
When the language used was not in English.
When the SI was estimated in the context of nosocomial transmission.
Statistical analysis
As the studies differed in their temporality and location, i.e. SI could vary over time depending on the control measures implemented or the adaptive behaviour of populations as the epidemic spread. Therefore, heterogeneity in the mean of SI was expected. Moreover, studies investigated case-to-case intervals between primary and secondary cases with different designs or various methodological approaches. For these reasons, a random effects model with restricted maximum-likelihood (REML) estimator method was used. Heterogeneity with Cochran’s Q and the I2 tests was checked. In order to detect a publication bias, a funnel plot was performed with an Egger’s test. The meta-analysis was performed using R 3.6.1. software with the package metafor [18].
Sub-groups analysis
A sensitivity analysis based on the following subgroups was performed:
Statistical distribution of the SI as it could bias the reproductive number [14].
SI estimate of published versus re-estimated from raw data when SI statistics were lacking, especially for statistical distribution.
Country by supposing they could differ in organisation of their public health systems.
SI estimated with or without negative values could bias both the mean and the standard deviation.
Measures of control implemented or not during the study could have influenced behaviour of populations and transmission of the virus.
The meta-analysis was carried out on the basis of studies which reported the mean of the SI with its standard deviation (SD) calculated from the number of infector/infected pairs. In case the mean and/or SD and/or distribution of SI was not directly reported in the publication, raw data was used. Therefore, 15 SI from 14 studies were re-estimated as following: if the statistical distribution reported in the publication was gamma, the mean of SI was αβ and its standard-deviation 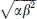 with α the shape and β the scale as the parameters of the distribution. If the reported distribution was a Weibull one, the mean and the SD were estimated from the mixdist package and the weibullparinv command of R. When the distribution was not known, the SI data was fitted according to four distributions, normal, lognormal, Weibull and gamma, and the best fit was chosen using AIC. Table 1 depicts the values re-estimated [19–32].
with α the shape and β the scale as the parameters of the distribution. If the reported distribution was a Weibull one, the mean and the SD were estimated from the mixdist package and the weibullparinv command of R. When the distribution was not known, the SI data was fitted according to four distributions, normal, lognormal, Weibull and gamma, and the best fit was chosen using AIC. Table 1 depicts the values re-estimated [19–32].
Statistics of SI re-estimated (in bold) from raw data of publications that do not or partly provide mean and/or SD and/or statistical distribution.
Results
The query retrieved 244 studies from PubMed. The 98 publications excluded concerned mainly clinical studies (51), epidemiological studies like seroprevalence (24) or meta-analysis/review (17). Among the 146 publications screened, 29 were excluded as their data were not sufficient for 22, and seven concerned the generation time only. The remaining 117 studies were assessed and 47 were excluded as they used previously published estimates of SI, mainly for modelling purpose. Finally, 67 studies and 76 estimates of SI were retained (Fig. 1). From the references of the eligible publications or meta-analysis/reviews, 13 additional studies with 14 SI estimates were found and included in the meta-analysis. A total of 90 SI estimates from 80 studies were obtained. Seventy five were retrieved directly from 66 publications and 15 were recalculated using raw data as described [10, 33–97].
 |
Figure 1 Cascade selection of publications reporting mean and standard-deviation for serial interval. SI: Serial interval. |
The SI estimates provided were mainly from studies conducted in Asian countries, 72.2%, mainly China and South Korea, 35.6 and 13.3% respectively. The SI estimates were retrieved mainly for alpha variant, 83.3%. A gamma distribution or a normal distribution were each used one out of three times to fit the SI (Tab. 2).
Characteristics description of the 90 SI estimates.
The overall estimate of SI was highest for the alpha variant, then the delta and omicron variants, respectively 5.17, 4.07, and 3.06 days (Tab. 3, Figs. 2, 4, and 5). The omicron SI estimate was lower than that of the alpha (Figs. 2 and 5). For all the variants, there was a heterogeneity according to the I2 and the Cochran tests. No publication bias were shown for alpha and omicron SI estimates contrary to delta (Figs. 3, 4, and 5).
 |
Figure 2 Forest plot of the 75 estimates of alpha variant’s SI included in the meta-analysis. SI: serial interval; SD: standard deviation; N pairs: number of pairs; RE model: random effect model; I2: I-square or index of heterogeneity statistic; Q: Cochran’s Q heterogeneity statistic. |
 |
Figure 3 Funnel plot of the 75 estimates of serial interval included in the meta-analysis for alpha variant. SI: serial interval. |
 |
Figure 4 Forest (a) and funnel (b) plot of the eight estimates of delta variant’s SI included in the meta-analysis. SI: serial interval; SD: standard deviation; N pairs: number of pairs; RE model: random effect model; I2: I-square or index of heterogeneity statistic; Q: Cochran’s Q heterogeneity statistic. |
 |
Figure 5 Forest (a) and funnel (b) plot of the eight estimates of omicron variant’s SI included in the meta-analysis. SI: serial interval; SD: standard deviation; N pairs: number of pairs; RE model: random effect model; I2: I-square or index of heterogeneity statistic; Q: Cochran’s Q heterogeneity statistic. |
Meta-analysis for SI of three variants of concern and their summary statistics.
From the sub-group analysis only performed for alpha variant, the highest SI were found for studies conducted in China, for those the SI values fitted with a gamma distribution, and for SI estimate with no negative value. In all subgroups, heterogeneity remained significant. For the majority of estimates, there was no publication bias (Tab. 4).
Meta-analysis according to subgroups for alpha variant.
Discussion
The meta-analysis was unable to provide a suitable estimate of SI for COVID-19 modelling purposes although its uncertainty was reduced. The SI estimate for alpha variant was close to that found in Rai’s meta-analysis, 5.19 days (95% CI = 4.37 – 6.02), and lower than those found by Zhang et al. and Hussein et al., 5.35 (95% CI = 4.63 – 6.07) and 5.45 days (95% CI = 4.23 – 6.66), respectively [15–17]. However, like the previously published meta-analyses, our estimate was hampered by strong heterogeneity between the studies. Indeed, their SI estimates varied between 2.4 and 8.7 days. The heterogeneity of the present meta-analysis was also high, I2 = 97.1% as the included studies differed in their design, geographic locations, their populations, and the stage of the epidemic.
A part of this heterogeneity could be also explained by potentially large proportions of asymptomatic or presymptomatic infected individuals as reported by some studies, between 5 and 89% [98, 99]. These last proportions are more often discussed as limitations of the study rather than clearly quantified in the studies.
Individual behaviours are also greatly modified during an epidemic, people protect themselves from infection and follow the control measures advised by governments. These are factors explaining why the SI changed during the pandemic as shown by Ali et al. [100]. This work shows the SI decreased from 7.8 to 2.6 days during one month of the pandemic with implementation of control measures such as the delay in prophylactic isolation. This also emphasizes the importance of prophylactic isolation. Indeed, as shown by a meta-analysis, the SI correlated positively with the delay of isolation which drastically decreased after the peak of epidemic [101]. The studies included in the present meta-analysis also measured the SI at different stages of the epidemic and in various populations like households, clusters, work place. These stages were generally not clearly described probably explaining why the SI was not significantly different between studies reporting the implementation of control measures and those that did not. It was not possible to perform a sensitivity analysis using this information. SI were also estimated according to different health information systems, which could also lead to a significant source of heterogeneity. Meta-analysis of studies with common and standardised methodology will be needed to obtain more reliable SI, for example, by sharing methodologic guidelines.
Given the differences between studies described above, a subgroup analysis was performed for the alpha variant. It was not possible to appreciably reduce the heterogeneity, especially taking into account the statistical distribution. However, it is noteworthy that SI following gamma and Weibull distributions were higher than that of the normal distribution. Therefore, the choice of the statistical distribution to fit the SI values of a given study should be taken into account. Indeed, the SI differs significantly, 4.5 days for the lognormal distribution, 5.2 days for the normal one and more than 5.4 days for the Weibull and gamma distributions.
This meta-analysis gave SI estimates for variants of concern, alpha, delta and omicron. The omicron variant was significantly shorter than that of alpha, delta SI had an intermediate value. These results are surprising as the delta variant R0 is 1.8 times that of the alpha variant and the omicron variant infectivity is 2.8 times that of delta variant, even if SI among unvaccinated delta variant was not found to be significant from that of unvaccinated non delta variant in one study [2, 7, 94]. These discrepancies can likely be explained by unvaccinated or “remaining to vaccinate” population either more exposed or more susceptible to new variants, a weaker vaccine effectiveness for delta and omicron variants, and epidemiological surveillance and contact tracing more efficient in their organisation and availability of screening tests throughout the epidemic.
Although the Egger’s test was non-significant except for SI estimate of delta variant, a selection bias of the studies cannot be excluded. It was impossible to use the SI from other publications either because of the language or the unavailability of sufficient information about SI.
Despite subgroup analysis, heterogeneity remained considerable. In a modelling context, this meta-analysis enables the use of SI values from 4.5 to 5.6 days according to the statistical distribution. After three years of the pandemic, surveillance and COVID tracing systems are working in routine and could provide SI from the field in real-time allowing a better estimate of the basic and efficient reproduction numbers. The advantage of this real-time monitoring of the SI would make it possible to follow the evolution of the reproduction rate of the infection, the choice and rapid assessment of the control measures implemented, and the effectiveness of the vaccination campaign.
Nomenclature of abbreviations
COVID-19: Coronavirus disease 2019
Distr.: Statistical distribution
I 2 : I-square or index of heterogeneity statistic
REML: Restricted Maximum-Likelihood Estimator
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2
Conflict of interest
Jean-François Jusot certifies that he has no financial conflict of interest in connection with this article. The author reports the following conflict of interest: Jean-François Jusot is Editorial Board member in Life Sciences-Medicine of 4open by EDP Sciences. This manuscript contains original material that has not previously been published.
References
- Choi JY, Smith DM (2021), SARS-CoV-2 variants of concern. Yonsei Med J 62, 11, 961–968. https://doi.org/10.3349/ymj.2021.62.11.961. [CrossRef] [PubMed] [Google Scholar]
- Aleem A, Akbar Samad AB, Slenker AK (2022), Emerging variants of SARS-CoV-2 and novel therapeutics against Coronavirus (COVID-19), in StatPearls, StatPearls Publishing. [Google Scholar]
- Gatto M, Bertuzzo E, Mari L, Miccoli S, Carraro L, Casagrandi R, Rinaldo A (2020), Spread and dynamics of the COVID-19 epidemic in Italy: effects of emergency containment measures. Proc Natl Acad Sci USA 117, 19, 10484–10491. https://doi.org/10.1073/pnas.2004978117. [CrossRef] [PubMed] [Google Scholar]
- Fang Y, Nie Y, Penny M (2020), Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: a data-driven analysis. J Med Virol 92, 6, 645–659. https://doi.org/10.1002/jmv.25750. [CrossRef] [PubMed] [Google Scholar]
- Jones JH, Hazel A, Almquist Z (2020), Transmission-dynamics models for the SARS Coronavirus-2. Am J Hum Biol Off J Hum Biol Counc 32, 5, e23512. https://doi.org/10.1002/ajhb.23512. [Google Scholar]
- Grillo Ardila EK, Santaella-Tenorio J, Guerrero R, Bravo LE (2020), Mathematical model and COVID-19. Colomb Medica Cali Colomb 51, 2, e4277. https://doi.org/10.25100/cm.v51i2.4277. [Google Scholar]
- Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J (2020), The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med 27, 2, taaa021. https://doi.org/10.1093/jtm/taaa021. [CrossRef] [PubMed] [Google Scholar]
- Peak CM, Kahn R, Grad YH, Childs LM, Li R, Lipsitch M, Buckee CO (2020), Individual quarantine versus active monitoring of contacts for the mitigation of COVID-19: a modelling study. Lancet Infect Dis 20, 9, 1025–1033. https://doi.org/10.1016/S1473-3099(20)30361-3. [CrossRef] [PubMed] [Google Scholar]
- Riccardo F, Ajelli M, Andrianou XD, Bella A, Del Manso M, Fabiani M, Bellino S, Boros S, Urdiales AM, Marziano V, Rota MC, Filia A, D’Ancona F, Siddu A, Punzo O, Trentini F, Guzzetta G, Poletti P, Stefanelli P, Castrucci MR, Ciervo A, Di Benedetto C, Tallon M, Piccioli A, Brusaferro S, Rezza G, Merler S, Pezzotti P, COVID-19 Working Group, (2020), Epidemiological characteristics of COVID-19 cases and estimates of the reproductive numbers 1 month into the epidemic, Italy, 28 January to 31 March 2020. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 25, 49, 2000790. https://doi.org/10.2807/1560-7917.ES.2020.25.49.2000790. [Google Scholar]
- Talmoudi K, Safer M, Letaief H, Hchaichi A, Harizi C, Dhaouadi S, Derouiche S, Bouaziz I, Gharbi D, Najar N, Osman M, Cherif I, Mlallekh R, Ben-Ayed O, Ayedi Y, Bouabid L, Bougatef S, Ben-Alaya NBÉ, Chahed MK (2020), Estimating transmission dynamics and serial interval of the first wave of COVID-19 infections under different control measures: a statistical analysis in Tunisia from February 29 to May 5, 2020. BMC Infect Dis 20, 1, 914. https://doi.org/10.1186/s12879-020-05577-4. [CrossRef] [PubMed] [Google Scholar]
- Vink MA, Bootsma MCJ, Wallinga J (2014), Serial intervals of respiratory infectious diseases: a systematic review and analysis. Am J Epidemiol 180, 9, 865–875. https://doi.org/10.1093/aje/kwu209. [CrossRef] [PubMed] [Google Scholar]
- Knight J, Mishra S (2020), Estimating effective reproduction number using generation time versus serial interval, with application to Covid-19 in the Greater Toronto Area, Canada. Infect Dis Model 5, 889–896. https://doi.org/10.1016/j.idm.2020.10.009. [PubMed] [Google Scholar]
- Ganyani T, Kremer C, Chen D, Torneri A, Faes C, Wallinga J, Hens N (2020), Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 25, 17, 2000257. https://doi.org/10.2807/1560-7917.ES.2020.25.17.2000257. [Google Scholar]
- Challen R, Brooks-Pollock E, Tsaneva-Atanasova K, Danon L (2021), Meta-analysis of the severe acute respiratory syndrome coronavirus 2 serial intervals and the impact of parameter uncertainty on the coronavirus disease 2019 reproduction number. Stat Methods Med Res. 2021 Dec 21:9622802211065159. Doi: https://doi.org/10.1177/09622802211065159. Epub ahead of print. [PubMed] [Google Scholar]
- Hussein M, Toraih E, Elshazli R, Fawzy M, Houghton A, Tatum D, Killackey M, Kandil E, Duchesne J (2021), Meta-analysis on Serial Intervals and Reproductive Rates for SARS-CoV-2. Ann Surg 273, 3, 416–423. https://doi.org/10.1097/SLA.0000000000004400. [CrossRef] [PubMed] [Google Scholar]
- Rai B, Shukla A, Dwivedi LK (2021), Estimates of serial interval for COVID-19: A systematic review and meta-analysis. Clin Epidemiol Glob Health 9, 157–161. https://doi.org/10.1016/j.cegh.2020.08.007. [CrossRef] [PubMed] [Google Scholar]
- Zhang P, Wang T, Xie SX (2020), Meta-analysis of several epidemic characteristics of COVID-19. J Data Sci JDS 18, 3, 536–549. https://doi.org/10.6339/JDS.202007_18(3).0019. [Google Scholar]
- R Core Team (2019), R: A language and environment for statistical computing, R Foundation for Statistical Computing, https://www.R-project.org/. [Google Scholar]
- Böhmer MM, Buchholz U, Corman VM, Hoch M, Katz K, Marosevic DV, Böhm S, Woudenberg T, Ackermann N, Konrad R, Eberle U, Treis B, Dangel A, Bengs K, Fingerle V, Berger A, Hörmansdorfer S, Ippisch S, Wicklein B, Grahl A, Pörtner K, Muller N, Zeitlmann N, Boender TS, Cai W, Reich A, An der Heiden M, Rexroth U, Hamouda O, Schneider J, Veith T, Mühlemann B, Wölfel R, Antwerpen M, Walter M, Protzer U, Liebl B, Haas W, Sing A, Drosten C, Zapf A (2020), Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect Dis 20, 8, 920–928. https://doi.org/10.1016/S1473-3099(20)30314-5. [CrossRef] [PubMed] [Google Scholar]
- Cereda D, Manica M, Tirani M, Rovida F, Demicheli V, Ajelli M, Poletti P, Trentini F, Guzzetta G, Marziano V, Piccarreta R, Barone A, Magoni M, Deandrea S, Diurno G, Lombardo M, Faccini M, Pan A, Bruno R, Pariani E, Grasselli G, Piatti A, Gramegna M, Baldanti F, Melegaro A, Merler S (2021), The early phase of the COVID-19 epidemic in Lombardy, Italy. Epidemics 37, 100528. https://doi.org/10.1016/j.epidem.2021.100528. [CrossRef] [PubMed] [Google Scholar]
- He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM (2020), Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26, 5, 672–675. https://doi.org/10.1038/s41591-020-0869-5. [CrossRef] [PubMed] [Google Scholar]
- Huang L, Zhang X, Zhang X, Wei Z, Zhang L, Xu J, Liang P, Xu Y, Zhang C, Xu A (2020), Rapid asymptomatic transmission of COVID-19 during the incubation period demonstrating strong infectivity in a cluster of youngsters aged 16–23 years outside Wuhan and characteristics of young patients with COVID-19: A prospective contact-tracing study. J Infect 80, 6, e1–e13. https://doi.org/10.1016/j.jinf.2020.03.006. [CrossRef] [Google Scholar]
- Ki M (2020), Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Korea. Epidemiol Health 42, e2020007. https://doi.org/10.4178/epih.e2020007. [CrossRef] [PubMed] [Google Scholar]
- Liao J, Fan S, Chen J, Wu J, Xu S, Guo Y, Li C, Zhang X, Wu C, Mou H, Song C, Li F, Wu G, Zhang J, Guo L, Liu H, Lv J, Xu L, Lang C (2020), Epidemiological and clinical characteristics of COVID-19 in adolescents and young adults. The Innovation 1, 1, 100001. https://doi.org/10.1016/j.xinn.2020.04.001. [CrossRef] [Google Scholar]
- Lu QB, Zhang Y, Liu MJ, Zhang H-Y, Jalali N, Zhang A-R, Li JC, Zhao H, Song QQ, Zhao TS, Zhao J, Liu HY, Du J, Teng AY, Zhou ZW, Zhou SX, Che TL, Wang T, Yang T, Guan XG, Peng XF, Wang YN, Zhang YY, Lv SM, Liu BC, Shi WQ, Zhang XA, Duan XG, Liu W, Yang Y, Fang LQ (2020), Epidemiological parameters of COVID-19 and its implication for infectivity among patients in China, 1 January to 11 February 2020. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 25, 40, 2000250. https://doi.org/10.2807/1560-7917.ES.2020.25.40.2000250. [Google Scholar]
- Mettler SK, Kim J, Maathuis MH (2020), Diagnostic serial interval as a novel indicator for contact tracing effectiveness exemplified with the SARS-CoV-2/COVID-19 outbreak in South Korea. Int J Infect Dis IJID Off Publ Int Soc Infect Dis 99, 346–351. https://doi.org/10.1016/j.ijid.2020.07.068. [Google Scholar]
- Tindale LC, Stockdale JE, Coombe M, Garlock ES, Lau WYV, Saraswat M, Zhang L, Chen D, Wallinga J, Colijn C (2020), Evidence for transmission of COVID-19 prior to symptom onset. eLife 9, e57149. https://doi.org/10.7554/eLife.57149. [CrossRef] [PubMed] [Google Scholar]
- Wang X, Pan Y, Zhang D, Chen L, Jia L, Li X, Yang P, Wang Q, Macintyre CR (2020), Basic epidemiological parameter values from data of real-world in mega-cities: the characteristics of COVID-19 in Beijing, China. BMC Infect Dis 20, 1, 526. https://doi.org/10.1186/s12879-020-05251-9. [CrossRef] [PubMed] [Google Scholar]
- Zhang J, Litvinova M, Wang W, Wang Y, Deng X, Chen X, Li M, Zheng W, Yi L, Chen X, Wu Q, Liang Y, Wang X, Yang J, Sun K, Longini IM Jr, Halloran ME, Wu P, Cowling BJ, Merler S, Viboud C, Vespignani A, Ajelli M, Yu H (2020), Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside Hubei province, China: a descriptive and modelling study. Lancet Infect Dis 20, 7, 793–802. https://doi.org/10.1016/S1473-3099(20)30230-9. [CrossRef] [PubMed] [Google Scholar]
- de Laval F, Grosset-Janin A, Delon F, Allonneau A, Tong C, Letois F, Couderc A, Sanchez MA, Destanque C, Biot F, Raynaud F, Bigaillon C, Ferraris O, Simon-Loriere E, Enouf V, Andriamanantena D, de Santi VP, Javelle E, Mérens A (2021), Lessons learned from the investigation of a COVID-19 cluster in Creil, France: effectiveness of targeting symptomatic cases and conducting contact tracing around them. BMC Infect Dis 21, 1, 457. https://doi.org/10.1186/s12879-021-06166-9. [CrossRef] [PubMed] [Google Scholar]
- Pung R, Mak TM, Kucharski AJ, Lee VJ (2021), Serial intervals in SARS-CoV-2 B.1.617.2 variant cases. The Lancet 398, 10303, 837–838. https://doi.org/10.1016/S0140-6736(21)01697-4. [CrossRef] [Google Scholar]
- Zhu W, Zhang M, Pan J, Yao Y, Wang W (2021), Effects of prolonged incubation period and centralized quarantine on the COVID-19 outbreak in Shijiazhuang, China: a modeling study. BMC Med 19, 1, 308. https://doi.org/10.1186/s12916-021-02178-z. [CrossRef] [PubMed] [Google Scholar]
- Aghaali M, Kolifarhood G, Nikbakht R, Saadati HM, Hashemi Nazari SS (2020), Estimation of the serial interval and basic reproduction number of COVID-19 in Qom, Iran, and three other countries: a data-driven analysis in the early phase of the outbreak. Transbound Emerg Dis 67, 6, 2860–2868. https://doi.org/10.1111/tbed.13656. [CrossRef] [PubMed] [Google Scholar]
- Ali ST, Wang L, Lau EHY, Xu X-K, Du Z, Wu Y, Leung GM, Cowling BJ (2020), Evolution of effective serial interval of SARS-CoV-2 by non-pharmaceutical interventions. Res Sq [Preprint]. 2020 Jun 1:rs.3.rs-32486. Doi: https://doi.org/10.21203/rs.3.rs-32486/v1. Update in: Science. 2020 Jul 21. [Google Scholar]
- Bae S, Kim H, Jung TY, Lim JA, Jo DH, Kang GS, Jeong SH, Choi DK, Kim HJ, Cheon YH, Chun MK, Kim M, Choi S, Chun C, Shin SH, Kim HK, Park YJ, Park O, Kwon HJ (2020), Epidemiological Characteristics of COVID-19 Outbreak at Fitness Centers in Cheonan, Korea. J Korean Med Sci 3531, e288. https://doi.org/10.3346/jkms.2020.35.e288. [CrossRef] [PubMed] [Google Scholar]
- Bi Q, Wu Y, Mei S, Ye C, Zou X, Zhang Z, Liu X, Wei L, Truelove SA, Zhang T, Gao W, Cheng C, Tang X, Wu X, Wu Y, Sun B, Huang S, Sun Y, Zhang J, Ma T, Lessler J, Feng T (2020), Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis 20, 8, 911–919. https://doi.org/10.1016/S1473-3099(20)30287-5. [CrossRef] [PubMed] [Google Scholar]
- Bui L, Thanh TN, Ngoc HN (2020), Early estimation of reproduction number of Covid-19 in Vietnam. medRxiv 2020.03.28.20046136. Doi: https://doi.org/10.1101/2020.03.28.20046136.. . [Google Scholar]
- Böhm S, Woudenberg T, Chen D, Marosevic DV, Böhmer MM, Hansen L, Wallinga J, Sing A, Katz K (2021), Epidemiology and transmission characteristics of early COVID-19 cases, 20 January–19 March 2020, in Bavaria, Germany. Epidemiol Infect 149, e65. https://doi.org/10.1017/S0950268821000510. [CrossRef] [PubMed] [Google Scholar]
- Chan YWD, Flasche S, Lam TLT, Leung M-HJ, Wong M-L, Lam H-Y, Chuang SK (2020), Transmission dynamics, serial interval and epidemiology of COVID-19 diseases in Hong Kong under different control measures. Wellcome Open Res 5, 91. https://doi.org/10.12688/wellcomeopenres.15896.1. [CrossRef] [Google Scholar]
- Chun JY, Baek G, Kim Y (2020), Transmission onset distribution of COVID-19. Int J Infect Dis IJID Off Publ Int Soc Infect Dis 99, 403–407. https://doi.org/10.1016/j.ijid.2020.07.075. [Google Scholar]
- Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA (2020), Serial interval of COVID-19 among publicly reported confirmed cases. Emerg Infect Dis 26, 6, 1341–1343. https://doi.org/10.3201/eid2606.200357. [CrossRef] [PubMed] [Google Scholar]
- Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA (2020), COVID-19 serial interval estimates based on confirmed cases in public reports from 86 Chinese cities. medRxiv [Preprint]. 2020 Apr 27:2020.04.23.20075796. Doi: https://doi.org/10.1101/2020.04.23.20075796. [Google Scholar]
- Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dörner L, Parker M, Bonsall D, Fraser C (2020), Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 368, 6491, eabb6936. https://doi.org/10.1126/science.abb6936. [CrossRef] [PubMed] [Google Scholar]
- Geismar C, Fragaszy E, Nguyen V, Fong WLE, Shrotri M, Beale S, Rodger A, Lampos V, Byrne T, Kovar J, Navaratnam AMD, Patel P, Aldridge RW, Hayward A; Virus Watch Collaborative (2021), Household serial interval of COVID-19 and the effect of Variant B.1.1.7: analyses from prospective community cohort study (Virus Watch). Wellcome Open Res 6, 224. https://doi.org/10.12688/wellcomeopenres.16974.2 . [PubMed] [Google Scholar]
- Gouvea-Reis FA, Oliveira PD, Silva DCS, Borja LS, Percio J, Souza FS, Peterka C, Feres C, de Oliveira J, Sodré G, Dos Santos W, de Moraes C (2021), COVID-19 Outbreak in a Large Penitentiary Complex, April-June 2020. Brazil. Emerg Infect Dis 27, 3, 924–927. https://doi.org/10.3201/eid2703.204079. [CrossRef] [PubMed] [Google Scholar]
- Haddad N, Clapham HE, Abou Naja H, Saleh M, Farah Z, Ghosn N, Mrad P, Howard N (2021), Calculating the serial interval of SARS-CoV-2 in Lebanon using 2020 contact-tracing data. BMC Infect Dis 21, 1, 1053. https://doi.org/10.1186/s12879-021-06761-w. [CrossRef] [PubMed] [Google Scholar]
- Kwok KO, Wong VWY, Wei WI, Wong SYS, Tang JWT (2020), Epidemiological characteristics of the first 53 laboratory-confirmed cases of COVID-19 epidemic in Hong Kong, 13 February 2020. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 25, 16, 2000155. https://doi.org/10.2807/1560-7917.ES.2020.25.16.2000155. [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z (2020), Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382, 13, 1199–1207. https://doi.org/10.1056/NEJMoa2001316. [CrossRef] [PubMed] [Google Scholar]
- Li L, Han ZG, Qin PZ, Liu W-H, Yang Z, Chen Z-Q, Li K, Xie CJ, Ma Y, Wang H, Huang Y, Fan SJ, Yan ZL, Ou CQ, Luo L (2022), Transmission and containment of the SARS-CoV-2 Delta variant of concern in Guangzhou, China: A population-based study. PLoS Negl Trop Dis 16, 1, e0010048. https://doi.org/10.1371/journal.pntd.0010048. [CrossRef] [PubMed] [Google Scholar]
- Li M, Liu K, Song Y, Wang M, Wu J (2020), Serial interval and generation interval for imported and local infectors, respectively, estimated using reported contact-tracing data of COVID-19 in China. Front Public Health 8, 577431. https://doi.org/10.3389/fpubh.2020.577431. [Google Scholar]
- Liu T, Qi L, Yao M, Tian K, Lin M, Jiang H, Zeng M, Huang J (2020), Serial Interval and Reproductive Number of COVID-19 Among 116 Infector-infectee Pairs – Jingzhou City, Hubei Province, China, 2020. China CDC Wkly 2, 27, 491–495. https://doi.org/10.46234/ccdcw2020.118. [CrossRef] [PubMed] [Google Scholar]
- Liu JY, Chen TJ, Hwang SJ (2020), Analysis of community-acquired COVID-19 cases in Taiwan. J Chin Med Assoc JCMA 83, 12, 1087–1092. https://doi.org/10.1097/JCMA.0000000000000411. [CrossRef] [PubMed] [Google Scholar]
- Ma S, Zhang J, Zeng M, Yun Q, Guo W, Zheng Y, Zhao S, Wang MH, Yang Z (2020), Epidemiological parameters of COVID-19: case series study. J Med Internet Res 22, 10, e19994. https://doi.org/10.2196/19994. [CrossRef] [PubMed] [Google Scholar]
- Najafi F, Izadi N, Hashemi-Nazari SS, Khosravi-Shadmani F, Nikbakht R, Shakiba E (2020), Serial interval and time-varying reproduction number estimation for COVID-19 in western Iran. New Microbes New Infect 36, 100715. https://doi.org/10.1016/j.nmni.2020.100715. [CrossRef] [PubMed] [Google Scholar]
- McAloon CG, Wall P, Griffin J, Casey M, Barber A, Codd M, Gormley E, Butler F, McV Messam LL, Walsh C, Teljeur C, Smyth B, Nolan P, Green MJ, O’Grady L, Culhane K, Buckley C, Carroll C, Doyle S, Martin J, More SJ (2021), Estimation of the serial interval and proportion of pre-symptomatic transmission events of COVID − 19 in Ireland using contact tracing data. BMC Public Health 21, 1, 805. https://doi.org/10.1186/s12889-021-10868-9. [CrossRef] [PubMed] [Google Scholar]
- Nishiura H, Linton NM, Akhmetzhanov AR (2020), Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis IJID Off Publ Int Soc Infect Dis 93, 284–286. https://doi.org/10.1016/j.ijid.2020.02.060. [Google Scholar]
- Thai PQ, Rabaa MA, Luong DH, Tan DQ, Quang TD, Quach H-L, Hoang Thi NA, Dinh PC, Nghia ND, Tu TA, Quang N, Phuc TM, Chau V, Khanh NC, Anh DD, Duong TN, Thwaites G, van Doorn HR, Choisy M; OUCRU COVID-19 Research Group (2021), The First 100 Days of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Control in Vietnam. Clin Infect Dis 729, e334–e342. https://doi.org/10.1093/cid/ciaa1130. [CrossRef] [PubMed] [Google Scholar]
- Pung R, Cook AR, Chiew CJ, Clapham HE, Sun Y, Li Z, Dickens BL, Ma S, Mak K, Tan CC, Heng D, Chen MI, Lee VJ (2021), Effectiveness of containment measures against COVID-19 in Singapore: Implications for other national containment efforts. Epidemiol Camb Mass 32, 1, 79–86. https://doi.org/10.1097/EDE.0000000000001257. [CrossRef] [PubMed] [Google Scholar]
- Prete CA, Buss L, Dighe A, Porto VB, da Silva Candido D, Ghilardi F, Pybus OG, de Oliveira WK, Croda JHR, Sabino EC, Faria NR, Donnelly CA, Nascimento VH (2021), Serial interval distribution of SARS-CoV-2 infection in Brazil. J Travel Med 28, 2, taaa115. https://doi.org/10.1093/jtm/taaa115. [CrossRef] [PubMed] [Google Scholar]
- Rajendrakumar AL, Nair ATN, Nangia C, Chourasia PK, Chourasia MK, Syed MG, Nair AS, Nair AB, Koya MSF (2021), Epidemic Landscape and Forecasting of SARS-CoV-2 in India. J Epidemiol Glob Health 11, 1, 55–59. https://doi.org/10.2991/jegh.k.200823.001. [Google Scholar]
- Reed IG, Walker ES, Landguth EL (2021), SARS-CoV-2 Serial Interval Variation, Montana, USA, March 1–July 31, 2020. Emerg Infect Dis 27, 5, 1486–1491. https://doi.org/10.3201/eid2705.204663. [CrossRef] [PubMed] [Google Scholar]
- Ren X, Li Y, Yang X, Li Z, Cui J, Zhu A, Zhao H, Yu J, Nie T, Ren M, Dong S, Cheng Y, Chen Q, Chang Z, Sun J, Wang L, Feng L, Gao GF, Feng Z, Li Z (2021), Evidence for pre-symptomatic transmission of coronavirus disease 2019 (COVID-19) in China. Influenza Other Respir Viruses 15, 1, 19–26. https://doi.org/10.1111/irv.12787. [CrossRef] [PubMed] [Google Scholar]
- Ryu S, Ali ST, Jang C, Kim B, Cowling BJ (2020), Effect of nonpharmaceutical interventions on transmission of severe acute respiratory syndrome Coronavirus 2, South Korea, 2020. Emerg Infect Dis 26, 10, 2406–2410. https://doi.org/10.3201/eid2610.201886. [CrossRef] [PubMed] [Google Scholar]
- Ryu S, Kim D, Lim JS, Ali ST, Cowling BJ (2022), Serial interval and transmission dynamics during SARS-CoV-2 delta variant predominance, South Korea. Emerg Infect Dis 28, 2, 407–410. https://doi.org/10.3201/eid2802.211774. [CrossRef] [PubMed] [Google Scholar]
- Saurabh S, Verma MK, Gautam V, Kumar N, Goel AD, Gupta MK, Bhardwaj P, Misra S (2020), Transmission dynamics of the COVID-19 epidemic at the district level in India: prospective observational study. JMIR Public Health Surveill 6, 4, e22678. https://doi.org/10.2196/22678. [CrossRef] [PubMed] [Google Scholar]
- Shi P, Gao Y, Shen Y, Chen E, Chen H, Liu J, Chen Y, Xiao Y, Wang K, Shi C, Lu B (2021), Characteristics and evaluation of the effectiveness of monitoring and control measures for the first 69 Patients with COVID-19 from 18 January 2020 to 2 March in Wuxi, China. Sustain Cities Soc 64, 102559. https://doi.org/10.1016/j.scs.2020.102559. [CrossRef] [PubMed] [Google Scholar]
- Son H, Lee H, Lee M, Eun Y, Park K, Kim S, Park W, Kwon S, Ahn B, Kim D, Kim C (2020), Epidemiological characteristics of and containment measures for COVID-19 in Busan, Korea. Epidemiol Health 42, e2020035. https://doi.org/10.4178/epih.e2020035. [CrossRef] [PubMed] [Google Scholar]
- Song JS, Lee J, Kim M, Jeong HS, Kim MS, Kim SG, Yoo HN, Lee JJ, Lee HY, Lee SE, Kim EJ, Rhee JE, Kim IH, Park YJ (2022), Serial intervals and household transmission of SARS-CoV-2 Omicron Variant, South Korea, 2021. Emerg Infect Dis 283, 756–759. https://doi.org/10.3201/eid2803.212607. [CrossRef] [PubMed] [Google Scholar]
- Viego V, Geri M, Castiglia J, Jouglard E (2020), Incubation period and serial interval of Covid-19 in a chain of infections in Bahia Blanca (Argentina). Cienc Saude Coletiva 25, 9, 3503–3510. https://doi.org/10.1590/1413-81232020259.20852020. [CrossRef] [PubMed] [Google Scholar]
- Wang Y, Teunis P (2020), Strongly heterogeneous transmission of COVID-19 in Mainland China: local and regional variation. Front Med 7, 329. https://doi.org/10.3389/fmed.2020.00329. [CrossRef] [Google Scholar]
- Wang K, Zhao S, Liao Y, Zhao T, Wang X, Zhang X, Jiao H, Li H, Yin Y, Wang MH, Xiao L, Wang L, He D (2020), Estimating the serial interval of the novel coronavirus disease (COVID-19) based on the public surveillance data in Shenzhen, China, from 19 January to 22 February 2020. Transbound Emerg Dis 67, 6, 2818–2822. https://doi.org/10.1111/tbed.13647. [CrossRef] [PubMed] [Google Scholar]
- Wong J, Chaw L, Koh WC, Alikhan MF, Jamaludin SA, Poh WWP, Naing L (2020), Epidemiological investigation of the first 135 COVID-19 Cases in Brunei: implications for surveillance, control, and travel restrictions. Am J Trop Med Hyg 103, 4, 1608–1613. https://doi.org/10.4269/ajtmh.20-0771. [CrossRef] [PubMed] [Google Scholar]
- Wu JT, Leung K, Bushman M, Kishore N, Niehus R, de Salazar PM, Cowling BJ, Lipsitch M, Leung GM (2020), Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 26, 4, 506–510. https://doi.org/10.1038/s41591-020-0822-7. [CrossRef] [PubMed] [Google Scholar]
- Xu X, Liu X, Wang L, Ali ST, Du Z, Bosetti P, Cowling BJ, Wu Y (2020), Household transmissions of SARS-CoV-2 in the time of unprecedented travel lockdown in China. medRxiv [Preprint]. 2020 Mar 6:2020.03.02.20029868. Doi: https://doi.org/10.1101/2020.03.02.20029868.. [Google Scholar]
- Yang L, Dai J, Zhao J, Wang Y, Deng P, Wang J (2020), Estimation of incubation period and serial interval of COVID-19: analysis of 178 cases and 131 transmission chains in Hubei province. China. Epidemiol Infect 148, e117. https://doi.org/10.1017/S0950268820001338. [CrossRef] [PubMed] [Google Scholar]
- You C, Deng Y, Hu W, Sun J, Lin Q, Zhou F, Pang CH, Zhang Y, Chen Z, Zhou XH (2020), Estimation of the time-varying reproduction number of COVID-19 outbreak in China. Int J Hyg Environ Health 228, 113555. https://doi.org/10.1016/j.ijheh.2020.113555. [CrossRef] [PubMed] [Google Scholar]
- Quach HL, Nguyen KC, Hoang NA, Pham TQ, Tran DN, Le MTQ, Do HT, Vien CC, Phan LT, Ngu ND, Tran TA, Phung DC, Tran QD, Dang TQ, Dang DA, Vogt F (2021), Association of public health interventions and COVID-19 incidence in Vietnam, January to December 2020. Int J Infect Dis 110, S28–S43. https://doi.org/10.1016/j.ijid.2021.07.044. [CrossRef] [PubMed] [Google Scholar]
- Hong K, Yum S, Kim J, Chun BC (2020), The serial interval of COVID-19 in Korea: 1,567 pairs of symptomatic cases from contact tracing. J Korean Med Sci 35, 50, e435. https://doi.org/10.3346/jkms.2020.35.e435. [CrossRef] [PubMed] [Google Scholar]
- Zhang M, Xiao J, Deng A, Zhang Y, Zhuang Y, Hu T, Li J, Tu H, Li B, Zhou Y, Yuan J, Luo L, Liang Z, Huang Y, Ye G, Cai M, Li G, Yang B, Xu B, Huang X, Cui Y, Ren D, Zhang Y, Kang M, Li Y (2021), Transmission dynamics of an outbreak of the COVID-19 Delta Variant B.1.617.2 – Guangdong Province, China, May–June 2021. China CDC Wkly 3, 27, 584–586. https://doi.org/10.46234/ccdcw2021.148. [CrossRef] [PubMed] [Google Scholar]
- Zhao H, Zhang Z, Lun W, Chen Z, Lu X, Li J, Qiu F, Li S, Mao C, Lu Y, Chen J, He Q, Lu J, Yang Z (2021), Transmission dynamics and successful control measures of SARS-CoV-2 in the mega-size city of Guangzhou, China. Medicine (Baltimore) 100, 48, e27846. https://doi.org/10.1097/MD.0000000000027846. [CrossRef] [PubMed] [Google Scholar]
- Zhao S, Tang B, Musa SS, Ma S, Zhang J, Zeng M, Yun Q, Guo W, Zheng Y, Yang Z, Peng Z, Chong MK, Javanbakht M, He D, Wang MH (2021), Estimating the generation interval and inferring the latent period of COVID-19 from the contact tracing data. Epidemics 36, 100482. https://doi.org/10.1016/j.epidem.2021.100482. [CrossRef] [PubMed] [Google Scholar]
- Zhao S, Gao D, Zhuang Z, Chong MKC, Cai Y, Ran J, Cao P, Wang K, Lou Y, Wang W, Yang L, He D, Wang M (2020), Estimating the serial interval of the novel coronavirus disease (COVID-19): a statistical analysis using the public data in Hong Kong from January 16 to February 15, 2020. Front Phys 8, 347. https://doi.org/10.3389/fphy.2020.00347. [CrossRef] [Google Scholar]
- Bushman M, Worby C, Chang HH, Kraemer MUG, Hanage WP (2021), Transmission of SARS-CoV-2 before and after symptom onset: impact of nonpharmaceutical interventions in China. Eur J Epidemiol 36, 4, 429–439. https://doi.org/10.1007/s10654-021-00746-4. [CrossRef] [PubMed] [Google Scholar]
- Backer JA, Eggink D, Andeweg SP, Veldhuijzen IK, van Maarseveen N, Vermaas K, Vlaemynck B, Schepers R, van den Hof S, Reusken CB, Wallinga J (2022), Shorter serial intervals in SARS-CoV-2 cases with Omicron BA.1 variant compared with Delta variant, the Netherlands, 13 to 26 December 2021. Eurosurveillance 27, 6, 2200042. https://doi.org/10.2807/1560-7917.ES.2022.27.6.2200042. [CrossRef] [Google Scholar]
- Kim D, Ali ST, Kim S, Jo J, Lim JS, Lee S, Ryu S (2022), Estimation of serial interval and reproduction number to quantify the transmissibility of SARS-CoV-2 Omicron variant in South Korea. Viruses 14, 3, 533. https://doi.org/10.3390/v14030533. [CrossRef] [PubMed] [Google Scholar]
- Kremer C, Braeye T, Proesmans K, André E, Torneri A, Hens N (2022), Observed serial intervals of SARS-CoV-2 for the Omicron and Delta variants in Belgium based on contact tracing data, 19 November to 31 December 2021. Preprint. https://doi.org/10.1101/2022.01.28.22269756. [Google Scholar]
- Ratovoson R, Razafimahatratra R, Randriamanantsoa L, Raberahona M, Rabarison HJ, Rahaingovahoaka FN, Andriamasy EH, Herindrainy P, Razanajatovo N, Andriamandimby SF, Dussart P, Shoenhals M, Randria MJDD, Heraud JM, Randremanana RV (2022), Household transmission of COVID-19 among the earliest cases in Antananarivo, Madagascar. Influenza Other Respir Viruses 16, 1, 48–55. https://doi.org/10.1111/irv.12896. [CrossRef] [PubMed] [Google Scholar]
- Paireau J, Mailles A, Eisenhauer C, de Laval F, Delon F, Bosetti P, Salje H, Pontiès V, Cauchemez S (2022), Early chains of transmission of COVID-19 in France, January to March 2020. Eurosurveillance 27, 6, 2001953. https://doi.org/10.2807/1560-7917.ES.2022.27.6.2001953. [CrossRef] [Google Scholar]
- Ping K, Lei M, Gou Y, Tao Z, Yao G, Hu C, Tao Q, Zou Z, Wang D, Li S, Huang Y (2021), Epidemiologic characteristics of COVID-19 in Guizhou Province, China. J Infect Dev Ctries 15, 03, 389–397. https://doi.org/10.3855/jidc.12818. [CrossRef] [PubMed] [Google Scholar]
- Won YS, Kim JH, Ahn CY, Lee H (2021), Subcritical Transmission in the Early Stage of COVID-19 in Korea. Int J Environ Res Public Health 18, 3, 1265. https://doi.org/10.3390/ijerph18031265. [CrossRef] [PubMed] [Google Scholar]
- Kristiansen MF, Heimustovu BH, Borg SÁ, Mohr TH, Gislason H, Møller LF, Christiansen DH, Steig BÁ, Petersen MS, Strøm M, Gaini S (2021), Epidemiology and clinical course of first wave Coronavirus Disease Cases, Faroe Islands. Emerg Infect Dis 27, 3, 749–758. https://doi.org/10.3201/eid2703.202589. [CrossRef] [PubMed] [Google Scholar]
- Rahimi E, Hashemi Nazari SS, Mokhayeri Y, Sharhani A, Mohammadi R (2021), Nine-month trend of time-varying reproduction numbers of COVID-19 in West of Iran. J Res Health Sci 21, 2, e00517–e00517. https://doi.org/10.34172/jrhs.2021.54. [CrossRef] [PubMed] [Google Scholar]
- Hong K, Yum SJ, Kim JH, Chun BC (2021), Re-estimation of basic reproduction number of COVID-19 based on the epidemic curve by symptom onset date. Epidemiol Infect 149, e53. https://doi.org/10.1017/S0950268821000431. [CrossRef] [PubMed] [Google Scholar]
- Ogata T, Tanaka H, Irie F, Hirayama A, Takahashi Y (2022), Shorter incubation period among unvaccinated Delta Variant Coronavirus Disease 2019 patients in Japan. Int J Environ Res Public Health 19, 3, 1127. https://doi.org/10.3390/ijerph19031127. [CrossRef] [PubMed] [Google Scholar]
- Ji T, Chen HL, Xu J, Wu L-N, Li J-J, Chen K, Qin G (2020), Lockdown contained the spread of 2019 novel coronavirus disease in Huangshi City, China: early epidemiological findings. Clin Infect Dis 71, 6, 1454–1460. https://doi.org/10.1093/cid/ciaa390. [CrossRef] [PubMed] [Google Scholar]
- Xia W, Liao J, Li C, Li Y, Qian X, Sun X (2020), Transmission of Corona Virus Disease 2019 during the incubation period may lead to a quarantine loophole. Preprint. https://doi.org/10.1101/2020.03.06.20031955. [Google Scholar]
- Kwok KO, Wei WI, Huang Y, Kam KM, Chan EYY, Riley S, et al. (2021), Evolving epidemiological characteristics of COVID-19 in Hong Kong from January to August 2020: retrospective study. J Med Internet Res 23, 4, e26645. https://doi.org/10.2196/26645. [CrossRef] [PubMed] [Google Scholar]
- Park SY, Kim YM, Yi S, Lee S, Na B-J, Kim CB, et al. (2020), Coronavirus disease outbreak in call center, South Korea. Emerg Infect Dis 26, 8, 1666–1670. https://doi.org/10.3201/eid2608.201274. [CrossRef] [PubMed] [Google Scholar]
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. (2020), Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 382, 22, 2081–2090. https://doi.org/10.1056/NEJMoa2008457. [CrossRef] [PubMed] [Google Scholar]
- Ali ST, Wang L, Lau EHY, Xu X-K, Du Z, Wu Y, et al. (2020), Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science 369, 6507, 1106–1109. https://doi.org/10.1126/science.abc9004. [CrossRef] [PubMed] [Google Scholar]
- Ali ST, Yeung A, Shan S, Wang L, Gao H, Du Z, Xu XK, Wu P, Lau EHY, Cowling BJ (2022), Serial intervals and case isolation delays for Coronavirus Disease 2019: a systematic review and meta-analysis. Clin Infect Dis 74, 4, 685–694. https://doi.org/10.1093/cid/ciab491. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Jusot J-F. 2022. An update of serial interval estimates for COVID-19: a meta-analysis. 4open, 5, 16.
All Tables
Statistics of SI re-estimated (in bold) from raw data of publications that do not or partly provide mean and/or SD and/or statistical distribution.
All Figures
 |
Figure 1 Cascade selection of publications reporting mean and standard-deviation for serial interval. SI: Serial interval. |
| In the text | |
 |
Figure 2 Forest plot of the 75 estimates of alpha variant’s SI included in the meta-analysis. SI: serial interval; SD: standard deviation; N pairs: number of pairs; RE model: random effect model; I2: I-square or index of heterogeneity statistic; Q: Cochran’s Q heterogeneity statistic. |
| In the text | |
 |
Figure 3 Funnel plot of the 75 estimates of serial interval included in the meta-analysis for alpha variant. SI: serial interval. |
| In the text | |
 |
Figure 4 Forest (a) and funnel (b) plot of the eight estimates of delta variant’s SI included in the meta-analysis. SI: serial interval; SD: standard deviation; N pairs: number of pairs; RE model: random effect model; I2: I-square or index of heterogeneity statistic; Q: Cochran’s Q heterogeneity statistic. |
| In the text | |
 |
Figure 5 Forest (a) and funnel (b) plot of the eight estimates of omicron variant’s SI included in the meta-analysis. SI: serial interval; SD: standard deviation; N pairs: number of pairs; RE model: random effect model; I2: I-square or index of heterogeneity statistic; Q: Cochran’s Q heterogeneity statistic. |
| In the text | |

